Adding NIL sequence data to lab site¶
This section is for adding genomic sequencing data (.tsv files) onto the existing dataset provided and displayed on the NILs browser page on Andersenlab.org.
Basic Commands To Know Prior¶
You should freshen up on the following terminal commands.
- cd - change directories
- rm - delete files
- cp - make a copy
- git
Your Sequencing Data¶
You will use the gt_hmm_fill.tsv file output from the nil-ril-nf pipeline for this step.
Important
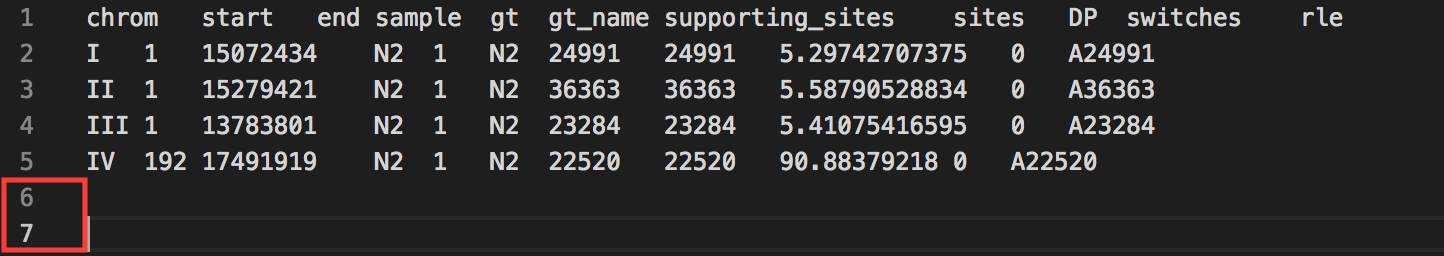
In order for your sequencing data to be properly added, it is important to make sure that there are no empty/additional lines located at the bottom of your .tsv file.
What you do not want
What you do want
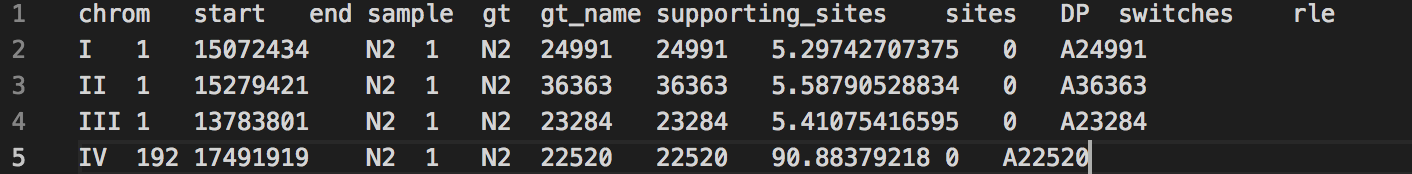
Once your file has no empty lines at the bottom, save the file and move onto the next instructions below.
Step By Step Instructions¶
- Clone the
andersenlab.orgrepo
git clone https://github.com/AndersenLab/andersenlab.github.io.git
-
[Optional] Create a new branch. This is a good idea if you are newer to github and want to make sure you don't break the lab website by doing something weird.
-
Add your .tsv file into the
pagesfolder of your Andersenlab github directory. -
Open terminal and use the
cdcommand to change directories into thepagesfolder in your Andersenlab github directory. If you did everything correctly, when you typelsinto your terminal, it should look something like this.

- Then run the following commands in your terminal (while still in your
pagesdirectory):
# create a new file called 'copy.tsv' with your data
cp yourFileName.tsv copy.tsv
# run python script which will add your data to full dataset
python addDataTogt_hmm.tsv.py
- After running the above commands, your sequencing data has now been added to the existing NILs dataset on Andersenlab.org. You can now remove your .tsv from the
pagesdirectory by using thermcommand in your terminal.
rm yourFileName.tsv
-
Finally, commit your changes and push your code to update the Andersenlab github.
-
[Optional] If you created a new branch, you need to merge this new branch back into the master branch for changes to take effect.
Viewing your results¶
Once all changes are pushed, you should be able to view your NIL genotypes in the NIL browser shiny app.